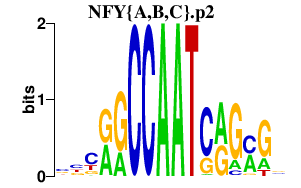
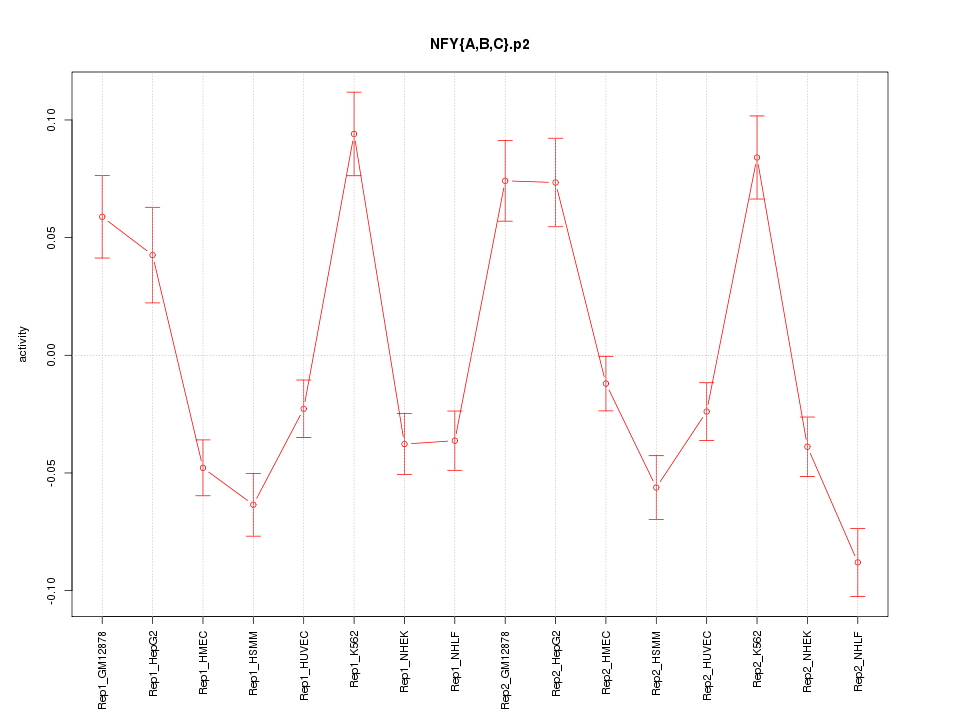
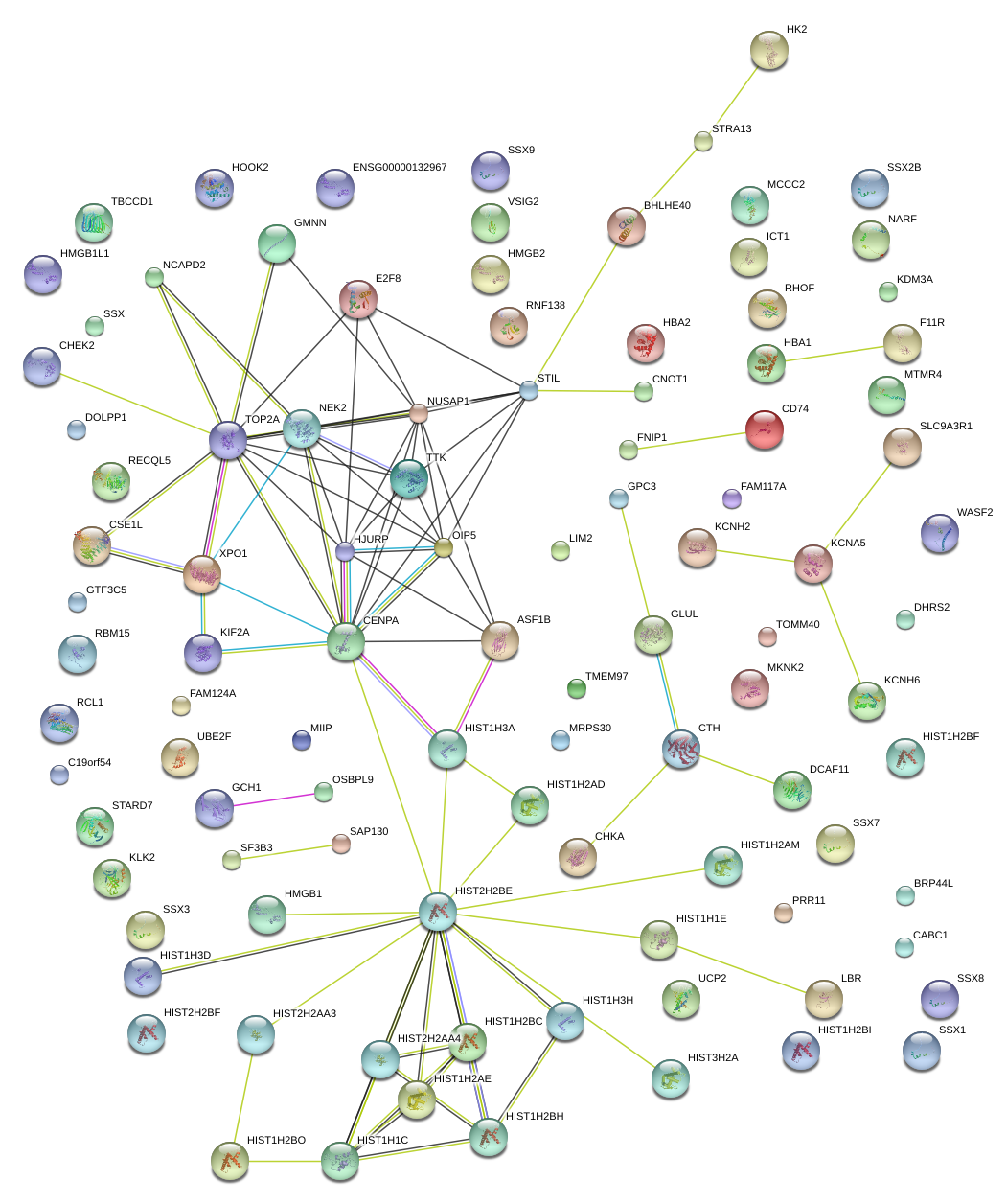
|
chr14_+_23175383
|
7.278
|
NM_005794
NM_182908
|
DHRS2
|
dehydrogenase/reductase (SDR family) member 2
|
|
chr17_+_78009895
|
5.489
|
|
NARF
|
nuclear prelamin A recognition factor
|
|
chr17_+_78009819
|
5.187
|
NM_001083608
NM_012336
NM_031968
|
NARF
|
nuclear prelamin A recognition factor
|
|
chr1_+_225194560
|
5.066
|
NM_020247
|
ADCK3
|
aarF domain containing kinase 3
|
|
chr1_+_225194582
|
4.959
|
|
ADCK3
|
aarF domain containing kinase 3
|
|
chr1_+_225194616
|
4.920
|
|
ADCK3
|
aarF domain containing kinase 3
|
|
chr17_-_35827568
|
4.794
|
NM_001067
|
TOP2A
|
topoisomerase (DNA) II alpha 170kDa
|
|
chr1_+_225194622
|
4.624
|
|
ADCK3
|
aarF domain containing kinase 3
|
|
chr17_+_23670247
|
4.515
|
NM_014573
|
TMEM97
|
transmembrane protein 97
|
|
chr17_-_53950191
|
4.499
|
NM_004687
|
MTMR4
|
myotubularin related protein 4
|
|
chr4_-_174492029
|
4.441
|
NM_001130688
NM_002129
|
HMGB2
|
high-mobility group box 2
|
|
chr19_-_14108400
|
4.357
|
NM_018154
|
ASF1B
|
ASF1 anti-silencing function 1 homolog B (S. cerevisiae)
|
|
chr14_-_54438913
|
4.196
|
NM_000161
NM_001024024
NM_001024070
NM_001024071
|
GCH1
|
GTP cyclohydrolase 1
|
|
chr17_+_54587641
|
4.052
|
NM_018304
|
PRR11
|
proline rich 11
|
|
chr1_+_110683586
|
3.829
|
|
RBM15
|
RNA binding motif protein 15
|
|
chr1_+_70649451
|
3.813
|
NM_001190463
NM_001902
NM_153742
|
CTH
|
cystathionase (cystathionine gamma-lyase)
|
|
chr1_-_147665783
|
3.806
|
|
HIST2H2BF
|
histone cluster 2, H2bf
|
|
chr1_+_110683580
|
3.806
|
|
RBM15
|
RNA binding motif protein 15
|
|
chrX_+_47999740
|
3.755
|
NM_005635
|
SSX1
|
synovial sarcoma, X breakpoint 1
|
|
chr2_+_74914742
|
3.669
|
|
HK2
|
hexokinase 2
|
|
chr11_-_67645433
|
3.665
|
NM_001277
NM_212469
|
CHKA
|
choline kinase alpha
|
|
chrX_-_132947141
|
3.515
|
|
GPC3
|
glypican 3
|
|
chr7_-_150305933
|
3.504
|
NM_000238
NM_172056
|
KCNH2
|
potassium voltage-gated channel, subfamily H (eag-related), member 2
|
|
chrX_+_52797004
|
3.480
|
NM_003147
NM_175698
NM_001164417
|
SSX2
SSX2B
|
synovial sarcoma, X breakpoint 2
synovial sarcoma, X breakpoint 2B
|
|
chrX_-_132947290
|
3.460
|
NM_001164617
NM_001164618
NM_001164619
NM_004484
|
GPC3
|
glypican 3
|
|
chr1_-_180627512
|
3.436
|
|
GLUL
|
glutamate-ammonia ligase
|
|
chr1_-_159257486
|
3.436
|
|
F11R
|
F11 receptor
|
|
chr1_-_27689129
|
3.418
|
|
WASF2
|
WAS protein family, member 2
|
|
chr17_+_78009336
|
3.393
|
NM_001038618
|
NARF
|
nuclear prelamin A recognition factor
|
|
chr19_-_14108267
|
3.380
|
|
ASF1B
|
ASF1 anti-silencing function 1 homolog B (S. cerevisiae)
|
|
chrX_-_52700674
|
3.354
|
NM_173358
|
SSX7
|
synovial sarcoma, X breakpoint 7
|
|
chr2_+_26862416
|
3.345
|
|
CENPA
|
centromere protein A
|
|
chr6_-_26164478
|
3.326
|
NM_005319
|
HIST1H1C
|
histone cluster 1, H1c
|
|
chr11_-_73371945
|
3.261
|
|
UCP2
|
uncoupling protein 2 (mitochondrial, proton carrier)
|
|
chr15_-_39412032
|
3.259
|
NM_007280
|
OIP5
|
Opa interacting protein 5
|
|
chr17_+_70256397
|
3.239
|
|
SLC9A3R1
|
solute carrier family 9 (sodium/hydrogen exchanger), member 3 regulator 1
|
|
chr17_-_45196516
|
3.231
|
NM_030802
|
FAM117A
|
family with sequence similarity 117, member A
|
|
chr17_+_16225130
|
3.227
|
|
|
|
|
chr17_-_77573974
|
3.207
|
NM_144998
|
STRA13
|
stimulated by retinoic acid 13 homolog (mouse)
|
|
chr15_-_20929608
|
3.138
|
|
HERC2P2
|
hect domain and RLD 2 pseudogene 2
|
|
chr17_-_45196465
|
3.062
|
|
FAM117A
|
family with sequence similarity 117, member A
|
|
chr12_+_6473552
|
3.015
|
NM_014865
|
NCAPD2
|
non-SMC condensin I complex, subunit D2
|
|
chr6_-_26305456
|
3.011
|
|
HIST1H3D
|
histone cluster 1, H3d
|
|
chr15_+_39412447
|
2.984
|
|
NUSAP1
|
nucleolar and spindle associated protein 1
|
|
chr2_-_96238296
|
2.959
|
NM_020151
|
STARD7
|
StAR-related lipid transfer (START) domain containing 7
|
|
chr9_+_130883199
|
2.954
|
NM_001135917
NM_020438
|
DOLPP1
|
dolichyl pyrophosphate phosphatase 1
|
|
chr6_+_80771108
|
2.893
|
|
TTK
|
TTK protein kinase
|
|
chr1_-_148080888
|
2.890
|
NM_001040874
NM_003516
|
HIST2H2AA4
HIST2H2AA3
|
histone cluster 2, H2aa4
histone cluster 2, H2aa3
|
|
chr16_+_162845
|
2.881
|
NM_000517
|
HBA1
HBA2
|
hemoglobin, alpha 1
hemoglobin, alpha 2
|
|
chr12_+_6473519
|
2.880
|
|
NCAPD2
|
non-SMC condensin I complex, subunit D2
|
|
chr16_-_57221235
|
2.875
|
|
CNOT1
|
CCR4-NOT transcription complex, subunit 1
|
|
chr17_+_70256389
|
2.857
|
|
SLC9A3R1
|
solute carrier family 9 (sodium/hydrogen exchanger), member 3 regulator 1
|
|
chr16_+_166678
|
2.834
|
NM_000558
|
HBA1
HBA2
|
hemoglobin, alpha 1
hemoglobin, alpha 2
|
|
chr11_-_67645246
|
2.829
|
|
CHKA
|
choline kinase alpha
|
|
chr1_-_159257567
|
2.813
|
|
F11R
|
F11 receptor
|
|
chr14_+_23654161
|
2.808
|
|
DCAF11
|
DDB1 and CUL4 associated factor 11
|
|
chr17_+_70256341
|
2.804
|
NM_004252
|
SLC9A3R1
|
solute carrier family 9 (sodium/hydrogen exchanger), member 3 regulator 1
|
|
chr6_+_26381122
|
2.802
|
NM_003525
|
HIST1H2BI
|
histone cluster 1, H2bi
|
|
chr2_+_238540361
|
2.791
|
|
UBE2F
|
ubiquitin-conjugating enzyme E2F (putative)
|
|
chr2_-_61618898
|
2.779
|
NM_003400
|
XPO1
|
exportin 1 (CRM1 homolog, yeast)
|
|
chr14_+_23654307
|
2.777
|
NM_181357
|
DCAF11
|
DDB1 and CUL4 associated factor 11
|
|
chr2_-_96238234
|
2.751
|
|
STARD7
|
StAR-related lipid transfer (START) domain containing 7
|
|
chr1_-_223682394
|
2.750
|
NM_002296
|
LBR
|
lamin B receptor
|
|
chr2_-_128501334
|
2.711
|
NM_001145928
NM_024545
|
SAP130
|
Sin3A-associated protein, 130kDa
|
|
chr1_-_223682324
|
2.668
|
|
LBR
|
lamin B receptor
|
|
chr12_-_120715937
|
2.659
|
NM_019034
|
RHOF
|
ras homolog gene family, member F (in filopodia)
|
|
chr11_-_19219000
|
2.643
|
NM_024680
|
E2F8
|
E2F transcription factor 8
|
|
chr20_+_47096189
|
2.607
|
NM_001316
|
CSE1L
|
CSE1 chromosome segregation 1-like (yeast)
|
|
chr3_-_187771025
|
2.552
|
NM_001134415
|
TBCCD1
|
TBCC domain containing 1
|
|
chr1_+_12002098
|
2.522
|
NM_021933
|
MIIP
|
migration and invasion inhibitory protein
|
|
chr13_-_29937966
|
2.508
|
|
HMGB1
HMGB1P6
|
high-mobility group box 1
high-mobility group box 1 pseudogene 6
|
|
chr6_+_80771040
|
2.504
|
NM_001166691
NM_003318
|
TTK
|
TTK protein kinase
|
|
chr3_-_9414122
|
2.503
|
|
LOC440944
|
hypothetical LOC440944
|
|
chr1_+_12002138
|
2.495
|
|
MIIP
|
migration and invasion inhibitory protein
|
|
chr5_-_149772563
|
2.483
|
|
CD74
|
CD74 molecule, major histocompatibility complex, class II invariant chain
|
|
chr1_-_209915443
|
2.474
|
NM_002497
|
NEK2
|
NIMA (never in mitosis gene a)-related kinase 2
|
|
chr19_-_45948211
|
2.447
|
|
C19orf54
|
chromosome 19 open reading frame 54
|
|
chr17_+_70256370
|
2.430
|
|
SLC9A3R1
|
solute carrier family 9 (sodium/hydrogen exchanger), member 3 regulator 1
|
|
chr5_-_131160541
|
2.407
|
|
FNIP1
|
folliculin interacting protein 1
|
|
chr5_+_44844724
|
2.389
|
NM_016640
|
MRPS30
|
mitochondrial ribosomal protein S30
|
|
chr5_+_70918856
|
2.389
|
NM_022132
|
MCCC2
|
methylcrotonoyl-CoA carboxylase 2 (beta)
|
|
chr16_-_57221240
|
2.380
|
NM_016284
NM_206999
|
CNOT1
|
CCR4-NOT transcription complex, subunit 1
|
|
chr1_+_51968046
|
2.377
|
NM_148904
NM_148905
NM_148907
|
OSBPL9
|
oxysterol binding protein-like 9
|
|
chr6_-_166716353
|
2.374
|
|
BRP44L
|
brain protein 44-like
|
|
chr22_-_27467776
|
2.351
|
NM_001005735
NM_007194
NM_145862
|
CHEK2
|
CHK2 checkpoint homolog (S. pombe)
|
|
chr19_-_1993022
|
2.346
|
|
MKNK2
|
MAP kinase interacting serine/threonine kinase 2
|
|
chr9_+_4782833
|
2.342
|
NM_005772
|
RCL1
|
RNA terminal phosphate cyclase-like 1
|
|
chr2_-_96237665
|
2.338
|
|
STARD7
|
StAR-related lipid transfer (START) domain containing 7
|
|
chr6_+_24883133
|
2.332
|
NM_015895
|
GMNN
|
geminin, DNA replication inhibitor
|
|
chr17_+_54587882
|
2.330
|
|
PRR11
|
proline rich 11
|
|
chr1_+_51968096
|
2.319
|
|
OSBPL9
|
oxysterol binding protein-like 9
|
|
chr14_+_23653848
|
2.314
|
|
DCAF11
|
DDB1 and CUL4 associated factor 11
|
|
chr5_+_70918888
|
2.312
|
|
MCCC2
|
methylcrotonoyl-CoA carboxylase 2 (beta)
|
|
chr9_+_134895882
|
2.312
|
NM_001122823
NM_012087
|
GTF3C5
|
general transcription factor IIIC, polypeptide 5, 63kDa
|
|
chr15_+_39412183
|
2.311
|
NM_001129897
NM_016359
NM_018454
|
NUSAP1
|
nucleolar and spindle associated protein 1
|
|
chr2_-_234427884
|
2.303
|
|
HJURP
|
Holliday junction recognition protein
|
|
chr14_+_23653745
|
2.298
|
NM_001163484
|
DCAF11
|
DDB1 and CUL4 associated factor 11
|
|
chr1_+_148089251
|
2.287
|
NM_001040874
NM_003516
|
HIST2H2AA4
HIST2H2AA3
|
histone cluster 2, H2aa4
histone cluster 2, H2aa3
|
|
chr19_+_50086350
|
2.283
|
|
TOMM40
|
translocase of outer mitochondrial membrane 40 homolog (yeast)
|
|
chr1_-_47552238
|
2.271
|
NM_001048166
NM_003035
|
STIL
|
SCL/TAL1 interrupting locus
|
|
chr2_+_26862368
|
2.271
|
NM_001809
NM_001042426
|
CENPA
|
centromere protein A
|
|
chr13_-_29938029
|
2.266
|
NM_002128
|
HMGB1
|
high-mobility group box 1
|
|
chr2_+_86521999
|
2.255
|
NM_001146688
|
KDM3A
|
lysine (K)-specific demethylase 3A
|
|
chr2_-_234427921
|
2.254
|
NM_018410
|
HJURP
|
Holliday junction recognition protein
|
|
chr9_+_134896194
|
2.251
|
|
GTF3C5
|
general transcription factor IIIC, polypeptide 5, 63kDa
|
|
chr5_+_162820094
|
2.249
|
NM_001142556
NM_001142557
NM_012484
NM_012485
|
HMMR
|
hyaluronan-mediated motility receptor (RHAMM)
|
|
chr1_+_110683467
|
2.249
|
NM_022768
|
RBM15
|
RNA binding motif protein 15
|
|
chr7_-_156495985
|
2.247
|
NM_005515
|
MNX1
|
motor neuron and pancreas homeobox 1
|
|
chr1_-_43945597
|
2.236
|
|
LOC100132774
|
hypothetical LOC100132774
|
|
chr5_+_70918893
|
2.235
|
|
MCCC2
|
methylcrotonoyl-CoA carboxylase 2 (beta)
|
|
chr5_-_149772469
|
2.233
|
NM_001025159
NM_004355
NM_001025158
|
CD74
|
CD74 molecule, major histocompatibility complex, class II invariant chain
|
|
chr14_+_23653819
|
2.231
|
NM_025230
|
DCAF11
|
DDB1 and CUL4 associated factor 11
|
|
chr10_-_69267774
|
2.222
|
NM_021800
NM_201262
|
DNAJC12
|
DnaJ (Hsp40) homolog, subfamily C, member 12
|
|
chr11_+_63710206
|
2.219
|
|
STIP1
|
stress-induced-phosphoprotein 1
|
|
chr9_-_6635564
|
2.184
|
NM_000170
|
GLDC
|
glycine dehydrogenase (decarboxylating)
|
|
chr6_+_32515596
|
2.167
|
NM_019111
|
HLA-DRA
|
major histocompatibility complex, class II, DR alpha
|
|
chr19_+_54962075
|
2.147
|
|
AP2A1
|
adaptor-related protein complex 2, alpha 1 subunit
|
|
chr6_-_31938530
|
2.143
|
|
NEU1
|
sialidase 1 (lysosomal sialidase)
|
|
chr1_-_195382189
|
2.135
|
NM_018136
|
ASPM
|
asp (abnormal spindle) homolog, microcephaly associated (Drosophila)
|
|
chr1_+_51968191
|
2.119
|
|
OSBPL9
|
oxysterol binding protein-like 9
|
|
chr5_+_70918935
|
2.117
|
|
MCCC2
|
methylcrotonoyl-CoA carboxylase 2 (beta)
|
|
chr1_-_28113540
|
2.116
|
|
RPA2
|
replication protein A2, 32kDa
|
|
chr20_-_45418818
|
2.106
|
NM_012408
NM_183047
NM_183048
|
ZMYND8
|
zinc finger, MYND-type containing 8
|
|
chr6_-_31938572
|
2.102
|
NM_000434
|
NEU1
|
sialidase 1 (lysosomal sialidase)
|
|
chr2_-_111152075
|
2.102
|
NM_004336
|
BUB1
|
budding uninhibited by benzimidazoles 1 homolog (yeast)
|
|
chr1_-_24067347
|
2.090
|
NM_000147
|
FUCA1
|
fucosidase, alpha-L- 1, tissue
|
|
chr2_+_10180304
|
2.086
|
NM_001034
|
RRM2
|
ribonucleotide reductase M2
|
|
chr1_+_210525490
|
2.066
|
NM_006243
|
PPP2R5A
|
protein phosphatase 2, regulatory subunit B', alpha
|
|
chr16_+_31792615
|
2.064
|
|
ZNF267
|
zinc finger protein 267
|
|
chr3_+_195336625
|
2.054
|
NM_005524
|
HES1
|
hairy and enhancer of split 1, (Drosophila)
|
|
chr19_-_45888259
|
2.053
|
NM_004756
|
NUMBL
|
numb homolog (Drosophila)-like
|
|
chr17_+_43373887
|
2.050
|
NM_018129
|
PNPO
|
pyridoxamine 5'-phosphate oxidase
|
|
chr8_+_6553278
|
2.048
|
NM_018361
|
AGPAT5
|
1-acylglycerol-3-phosphate O-acyltransferase 5 (lysophosphatidic acid acyltransferase, epsilon)
|
|
chr14_+_38714111
|
2.041
|
NM_002687
|
PNN
|
pinin, desmosome associated protein
|
|
chr3_-_187770790
|
2.029
|
|
TBCCD1
|
TBCC domain containing 1
|
|
chr2_+_86521939
|
2.026
|
|
KDM3A
|
lysine (K)-specific demethylase 3A
|
|
chr16_+_45281078
|
1.993
|
|
ORC6
PPA2
|
origin recognition complex, subunit 6
pyrophosphatase (inorganic) 2
|
|
chr1_+_148125090
|
1.992
|
NM_003517
|
HIST2H2AC
|
histone cluster 2, H2ac
|
|
chr11_+_63710242
|
1.989
|
|
STIP1
|
stress-induced-phosphoprotein 1
|
|
chr3_+_123036719
|
1.985
|
NM_018456
|
EAF2
|
ELL associated factor 2
|
|
chr7_+_137795387
|
1.985
|
NM_003852
NM_015905
|
TRIM24
|
tripartite motif containing 24
|
|
chr2_+_47483795
|
1.985
|
|
MSH2
|
mutS homolog 2, colon cancer, nonpolyposis type 1 (E. coli)
|
|
chr14_-_57834558
|
1.960
|
|
FLJ31306
|
hypothetical LOC379025
|
|
chr10_+_62208256
|
1.936
|
|
CDK1
|
cyclin-dependent kinase 1
|
|
chr16_+_56327168
|
1.933
|
|
KATNB1
|
katanin p80 (WD repeat containing) subunit B 1
|
|
chr3_-_52542748
|
1.924
|
NM_001134231
|
NT5DC2
|
5'-nucleotidase domain containing 2
|
|
chr20_+_60744231
|
1.912
|
NM_016354
|
SLCO4A1
|
solute carrier organic anion transporter family, member 4A1
|
|
chr10_+_62208094
|
1.904
|
NM_001170406
NM_001170407
NM_001786
NM_033379
|
CDK1
|
cyclin-dependent kinase 1
|
|
chr2_+_47483750
|
1.898
|
NM_000251
|
MSH2
|
mutS homolog 2, colon cancer, nonpolyposis type 1 (E. coli)
|
|
chr16_-_55042614
|
1.897
|
NM_007006
|
NUDT21
|
nudix (nucleoside diphosphate linked moiety X)-type motif 21
|
|
chr20_+_60744268
|
1.894
|
|
SLCO4A1
|
solute carrier organic anion transporter family, member 4A1
|
|
chr1_+_24158887
|
1.893
|
NM_017761
|
PNRC2
|
proline-rich nuclear receptor coactivator 2
|
|
chr17_-_73694669
|
1.883
|
NM_003258
|
TK1
|
thymidine kinase 1, soluble
|
|
chr1_+_24158898
|
1.881
|
|
PNRC2
|
proline-rich nuclear receptor coactivator 2
|
|
chr17_-_77261028
|
1.871
|
NM_001040025
|
ARL16
|
ADP-ribosylation factor-like 16
|
|
chr7_+_98908449
|
1.869
|
NM_001013258
NM_213603
|
ZNF789
|
zinc finger protein 789
|
|
chr19_-_17377383
|
1.863
|
NM_004335
|
BST2
|
bone marrow stromal cell antigen 2
|
|
chr11_+_63710162
|
1.848
|
NM_006819
|
STIP1
|
stress-induced-phosphoprotein 1
|
|
chr6_+_43005214
|
1.820
|
|
CNPY3
|
canopy 3 homolog (zebrafish)
|
|
chr8_-_145612636
|
1.804
|
NM_017767
|
SLC39A4
|
solute carrier family 39 (zinc transporter), member 4
|
|
chr15_+_26876480
|
1.800
|
|
|
|
|
chr9_+_4782876
|
1.798
|
|
RCL1
|
RNA terminal phosphate cyclase-like 1
|
|
chr6_+_37245860
|
1.796
|
NM_002648
|
PIM1
|
pim-1 oncogene
|
|
chr10_-_101180191
|
1.793
|
NM_002079
|
GOT1
|
glutamic-oxaloacetic transaminase 1, soluble (aspartate aminotransferase 1)
|
|
chr6_+_26307691
|
1.792
|
NM_003522
|
HIST1H2BF
|
histone cluster 1, H2bf
|
|
chr1_+_51855343
|
1.786
|
|
OSBPL9
|
oxysterol binding protein-like 9
|
|
chr3_+_9414290
|
1.779
|
NM_001080517
|
SETD5
|
SET domain containing 5
|
|
chr11_+_34417109
|
1.773
|
|
CAT
|
catalase
|
|
chr6_-_31890765
|
1.769
|
NM_005527
|
HSPA1L
|
heat shock 70kDa protein 1-like
|
|
chr7_-_150306169
|
1.748
|
|
KCNH2
|
potassium voltage-gated channel, subfamily H (eag-related), member 2
|
|
chr2_+_10180145
|
1.747
|
NM_001165931
|
RRM2
|
ribonucleotide reductase M2
|
|
chr22_-_40672493
|
1.735
|
|
CENPM
|
centromere protein M
|
|
chr19_+_50086362
|
1.732
|
|
TOMM40
|
translocase of outer mitochondrial membrane 40 homolog (yeast)
|
|
chr1_-_148126089
|
1.731
|
NM_175065
|
HIST2H2AB
HIST2H2BE
|
histone cluster 2, H2ab
histone cluster 2, H2be
|
|
chr2_+_162908869
|
1.711
|
|
GCA
|
grancalcin, EF-hand calcium binding protein
|
|
chr2_+_47483694
|
1.710
|
|
MSH2
|
mutS homolog 2, colon cancer, nonpolyposis type 1 (E. coli)
|
|
chr15_+_57184568
|
1.708
|
NM_004701
|
CCNB2
|
cyclin B2
|
|
chr2_-_61618849
|
1.706
|
|
XPO1
|
exportin 1 (CRM1 homolog, yeast)
|
|
chr1_-_148050534
|
1.706
|
NM_001024599
NM_001161334
|
HIST2H2BF
|
histone cluster 2, H2bf
|
|
chr2_-_61618871
|
1.706
|
|
XPO1
|
exportin 1 (CRM1 homolog, yeast)
|
|
chr12_-_26877189
|
1.705
|
NM_002223
|
ITPR2
|
inositol 1,4,5-triphosphate receptor, type 2
|
|
chr1_+_62674913
|
1.704
|
NM_003368
|
USP1
|
ubiquitin specific peptidase 1
|
|
chr1_-_109742020
|
1.690
|
NM_002959
|
SORT1
|
sortilin 1
|
|
chr1_-_19101603
|
1.687
|
NM_003748
NM_170726
|
ALDH4A1
|
aldehyde dehydrogenase 4 family, member A1
|
|
chr2_+_27128024
|
1.683
|
|
AGBL5
|
ATP/GTP binding protein-like 5
|
|
chr7_+_137795719
|
1.680
|
|
TRIM24
|
tripartite motif containing 24
|
|
chr8_-_145713959
|
1.676
|
NM_004260
|
RECQL4
|
RecQ protein-like 4
|
|
chrX_+_48127911
|
1.674
|
NM_001034832
NM_001040612
NM_005636
NM_175729
|
SSX4B
SSX4
|
synovial sarcoma, X breakpoint 4B
synovial sarcoma, X breakpoint 4
|
|
chr19_+_50086357
|
1.672
|
|
TOMM40
|
translocase of outer mitochondrial membrane 40 homolog (yeast)
|
|
chr16_+_31792578
|
1.669
|
NM_003414
|
ZNF267
|
zinc finger protein 267
|
|
chr3_+_195336631
|
1.666
|
|
HES1
|
hairy and enhancer of split 1, (Drosophila)
|
|
chr4_-_84254873
|
1.661
|
|
PLAC8
|
placenta-specific 8
|
|
chr6_+_43005126
|
1.651
|
|
CNPY3
|
canopy 3 homolog (zebrafish)
|
|
chr1_-_167721723
|
1.649
|
NM_006996
|
SLC19A2
|
solute carrier family 19 (thiamine transporter), member 2
|
|
chr6_-_42821815
|
1.640
|
NM_003192
|
TBCC
|
tubulin folding cofactor C
|
|
chr1_-_35431318
|
1.638
|
|
SFPQ
|
splicing factor proline/glutamine-rich
|
|
chr16_+_2419418
|
1.636
|
|
CCNF
|
cyclin F
|
|
chrX_-_48661226
|
1.635
|
NM_006875
|
PIM2
|
pim-2 oncogene
|
|
chr11_-_46824416
|
1.626
|
NM_001008938
NM_014756
|
CKAP5
|
cytoskeleton associated protein 5
|
|
chr1_+_148726537
|
1.624
|
NM_025150
|
TARS2
|
threonyl-tRNA synthetase 2, mitochondrial (putative)
|